Current Projects
Bridge Builder
Salt bridges are molecular interactions between charged amino acids which can impart stability to proteins or protein complexes. I am working on a python-based program which can identify putative mutation sites on input protein complexes. The goal is to introduce mutations such that the stability of a complex is enhanced. Currently, my algorithm uses the centroid side chain distance to efficiently identify putative mutations. I hope to incorporate machine learning to predict whether mutations impart a stabilizing effect to a complex.

Viral Assembly
Viral assembly is an essential step in the life cycle of all viruses. My work focuses on the assembly of Paramecium Bursaria Chlorella Virus 1 (PBCV-1). PBCV-1 (pictured on the right) is made up of thousands of proteins. My work is geared toward understanding how electrostatics influence the coordinated assembly of these units. To this end, I use the Poisson–Boltzmann solver, Delphi, to characterize electrostatic features on the surface of viral proteins.

Motor Protein Dynamics
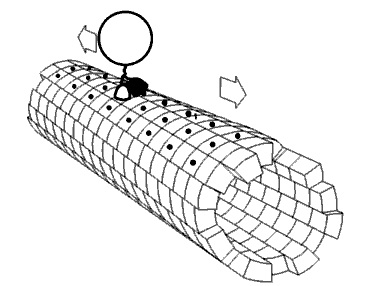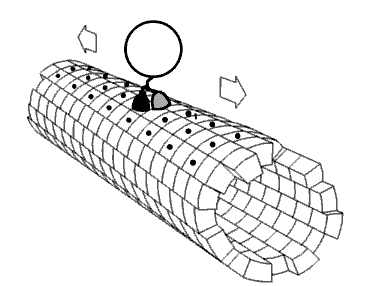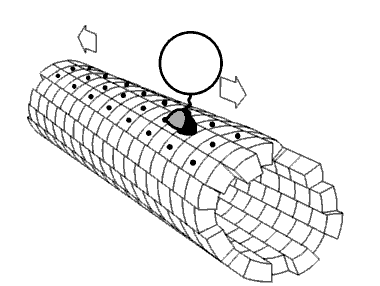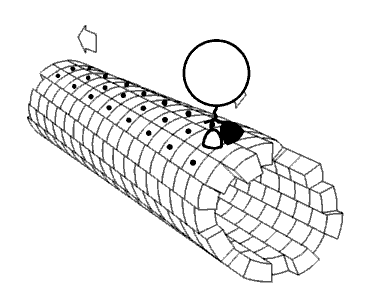
Molecular motors transport cellular cargo along protein fibers. My research focuses on a class of molecular motors known as kinesins. Kinesins move about microtubules and are involved in cellular processes such as mitosis. Because of this, microtubules and kinesins have been studied as potential anti-cancer drug targets.